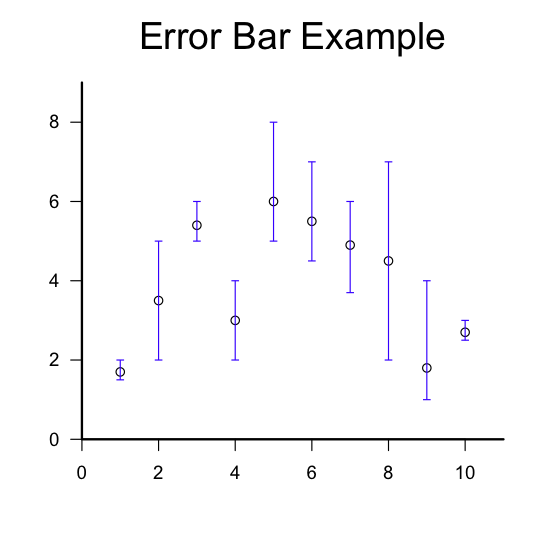
By Allan Roberts
This is a follow up to a previous post, Error Bars with R. The main difference is that the function used here allows the upper and lower limits of each error bar to be specified individually (rather than using a single parameter for the radius of an interval). Also, the “segments” function is used instead of the “arrows” function. The “segments” function is a lot like the “lines” function, except the endpoints of the line segments are given differently for the two functions. I’ve found that the segments function can be more useful if you wish to vectorize the drawing of multiple segments. (In the example below, the function “segments” is passed vectors of values; in general this is faster than using a for loop for the same purpose.)
Copying and pasting the script below should reproduce the example figure shown above. (Note: depending on whether you’re using a Mac or a PC, either the “quartz” or “windows” function should work to open a new graphics window, as indicated in the comments to the script.)
R Script
add.error.bars <- function(X,upper,lower,width,col=par( )$fg,lwd=1){
segments(X,lower,X,upper,col=col,lwd=lwd,lend=1);
segments(X-width/2,lower,X+width/2,lower,col=col,lwd=lwd,lend=1);
segments(X-width/2,upper,X+width/2,upper,col=col,lwd=lwd,lend=1);
}
X = 1:10; #A sequence of the integers from 1 to 10, inclusive.
Y = c(1.7, 3.5, 5.4,3,6,5.5,4.9,4.5,1.8,2.7); #Example values for y.
upper=c( 2, 5, 6, 4, 8, 7, 6, 7, 4, 3); #Upper limits for the error bars.
lower=c(1.5, 2, 5, 2, 5, 4.5, 3.7, 2, 1, 2.5); #Lower limits.
quartz(character(1),5,5); #Open a new graphics window on a Mac.
windows(character(1),5,5); # On a PC
plot.new();
par(usr=c(0,11, 0,max(upper)+1)); #Set the limits for the plotting window axes.
axis(1); #Draw the horizontal axis.
axis(2,las=1); #Draw the vertical axis.
box(bty=letters[12], lwd=2); #Draw a frame around the plot.
title(main=quote(Example),cex.main=2,font.main=1); #Add a title.
points(X,Y); #Plot the data points.
add.error.bars(X,lower,upper,width=0.2,col=rgb(0,0,1)); #Draw the error bars.